The PXMOD implementation differs from that described in [35] in the following points
Acquisition and Data Requirements
Image Data |
A dynamic PET data set with an neuroreceptor tracer which behaves kinetically similar to a 1-tissue compartment model. |
TAC 1 |
TAC from a receptor-rich region (such as basal ganglia for D2 receptors). |
TAC 2 |
TAC from a receptor-devoid region (such as cerebellum or frontal cortex for D2 receptors). |
Model Preprocessing
Two regional TACs (TAC1 and TAC2) are needed for Model Preprocessing.
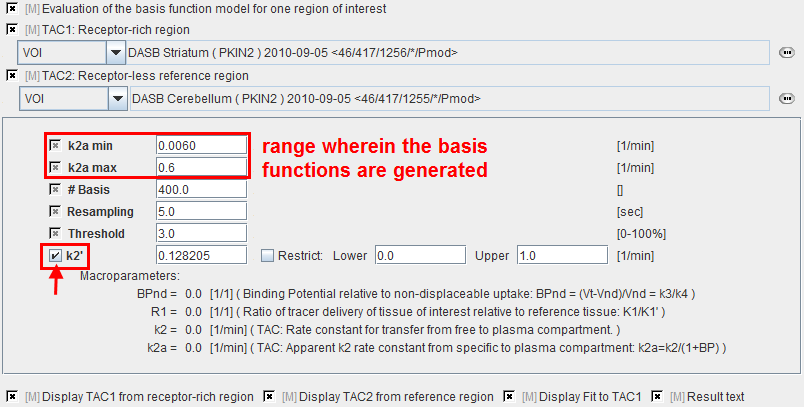
k2a min |
Minimal value of k2a (slowest decay of exponential). |
k2a max |
Maximal value of k2a (fastest decay of exponential). |
# Basis |
Number of basis functions between k2a min and k2a max. Note that increments are taken at logarithmic steps. This number is directly proportional to processing time. |
Resampling |
Specifies the interval of curve resampling which is required for performing the operation of exponential convolution. Resampling should be equal or smaller than the shortest frame duration. |
Threshold |
Discrimination threshold for background masking. |
k2' |
k2 of the reference tissue. It can be fixed at a specified value, or fitted. If it is checked, k2' is fitted using the SRTM method implemented in PKIN. This value will be used in pixel-wise SRTM2. |
BPnd |
Estimated binding potential (= k3/k4 according to the underlying model). |
R1 |
Ratio of tracer delivery in each pixel relative to the reference tissue (R1=K1/K1'). |
k2 |
Estimated rate constant k2. |
k2a |
k2a value which provides the best least squares fit in each voxel. |
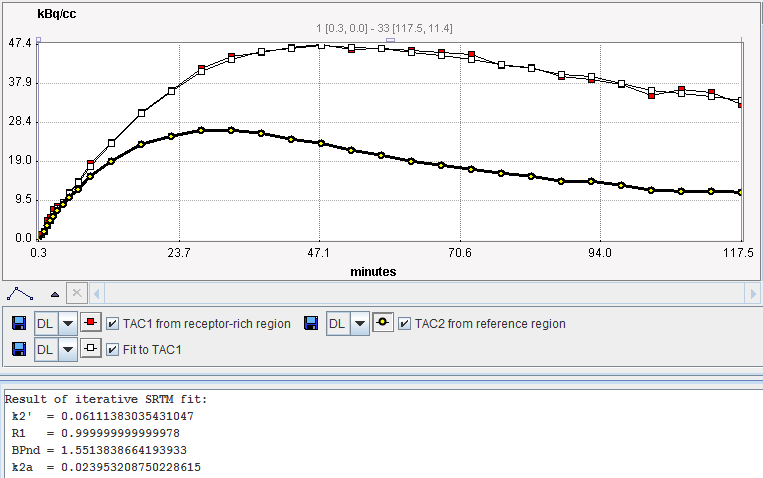
The result of the fit during Model Preprocessing is shown in the Result panel for inspection.
Map Parameters
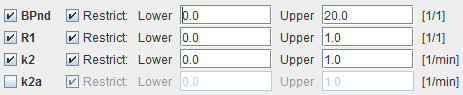
BPnd |
Estimated binding potential (BPnd= k3/k4 according to the underlying model). |
k2 |
Estimated efflux rate constant k2 . |
R1 |
Ratio of tracer delivery in each pixel relative to the reference tissue (R1=K1/K1'). Therefore the map often has a similar appearance to a perfusion image. |
k2a |
k2a value which provides the best least squares fit. |
Note: The k2a parametric map should be checked in the initial setup of a processing protocol. The estimated k2a values should not be truncated by too narrow k2a min and k2a max values.