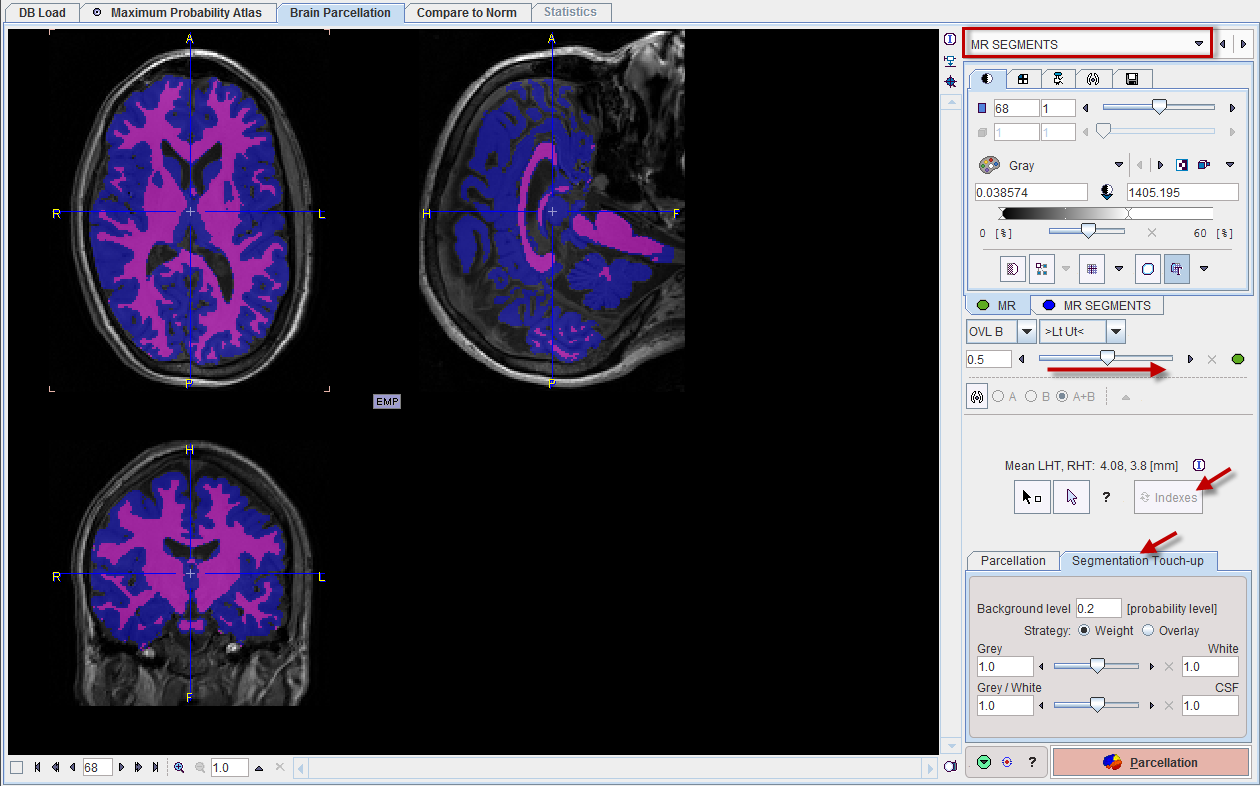
The result of the segmentation is shown as a fusion of the tissue segment map with the MR image on the MR SEGMENTS page. Note that the MR SEGMENTS image tab contains a label image with gray matter, white matter and CSF represented by the label values 1, 2 and 3, respectively.
Segmentation Touch-up
The label map is calculated from the probability maps of GM, WM and CSF. The relative extent of the tissue categories can be modified on the Segmentation Touch-up panel using two different methods. Any modification is immediately reflected in the display, but requires that the Indexes are recalculated before the parcellation can be started.
If the probability value in all of the GM, WM, and CSF maps is below the Background level, a pixel is classified as background and assigned the background label value of 0.
With the Weight strategy the following procedure is applied in all non-background pixels: The GM and WM probabilities are multiplied by the factor in the Grey/White field, and the CSF probability by the value in the CSF field. If the scaled probability of CSF is higher than the scaled GM and WM probabilities, the pixel is assigned the CSF label value of 3. Otherwise, a similar comparison is done between GM and WM: The GM probability is multiplied by the factor in the Grey field, and the WM probability by the value in the White field. If the scaled GM probability is higher than the scaled WM probability, the pixel is assigned the GM label value of 1, otherwise the WM label value of 2.
The Overlay strategy simply uses two thresholds for the GM and the WM probability map.
Please use the fusion slider to evaluate the segmentation quality. In case the result is not satisfactory, return to the previous page, modify the segmentation parameters, then activate Segment MR again.
Landmark Definition
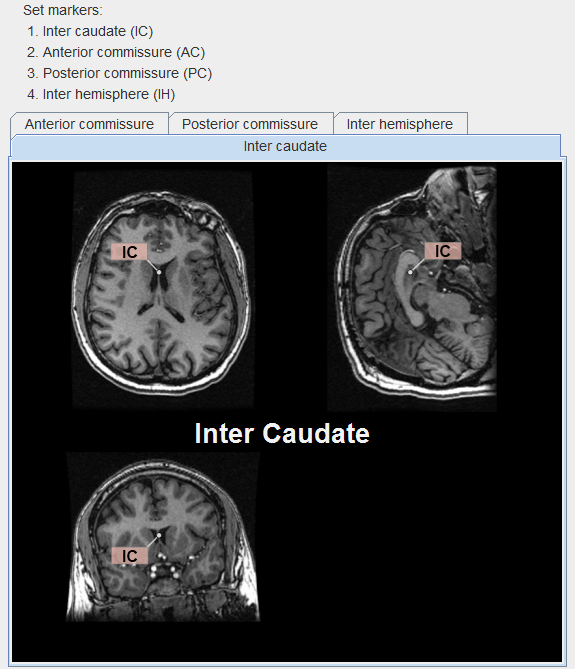
The parcellation procedure requires four anatomical landmarks
A visual help for this task can be opened with the ? button to the left of the Indexes button. It appears with the IC landmark on front as illustrated below.
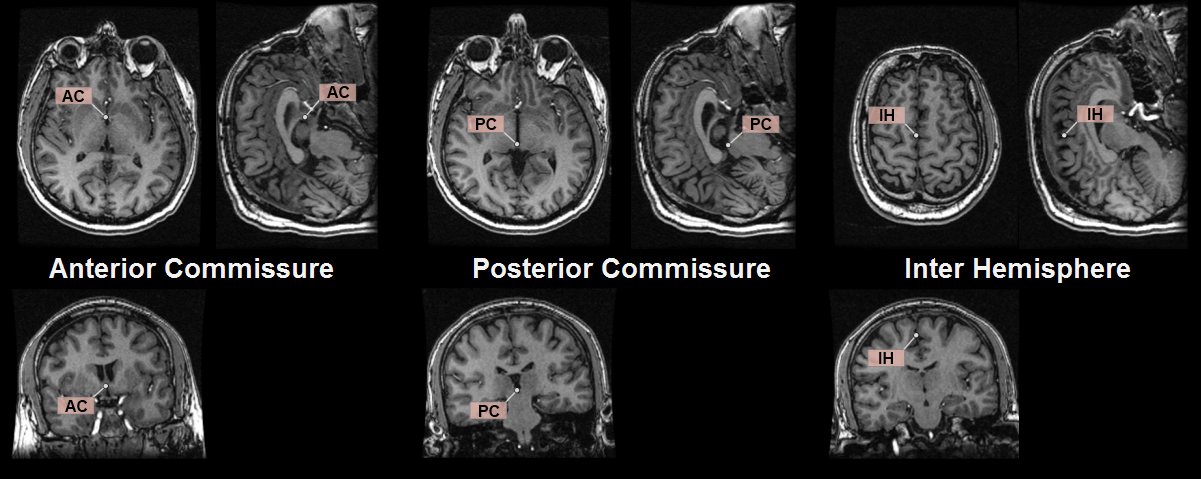
The location of the other three landmarks is illustrated below.
Estimates of the landmark locations are obtained as part of the segmentation. They can be inspected by activating the ![]() button. The Go to list allows to easily select any of the 4 landmarks, which is shown as a marker in the triangulated image. If the location is not accurate within a few millimeters, please adjust the position by dragging the marker. You may need to shift the fusion slider to the left in order to see the markers on top of only the MR.
button. The Go to list allows to easily select any of the 4 landmarks, which is shown as a marker in the triangulated image. If the location is not accurate within a few millimeters, please adjust the position by dragging the marker. You may need to shift the fusion slider to the left in order to see the markers on top of only the MR.
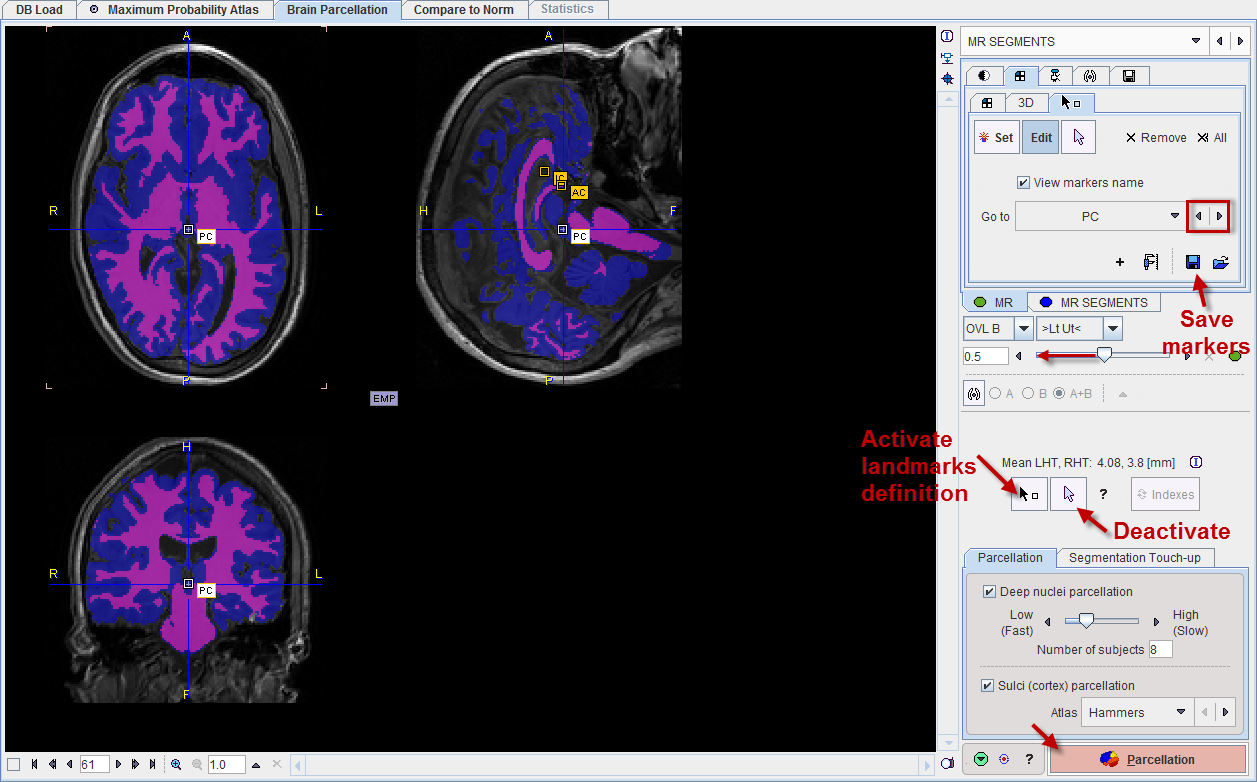
To avoid marker placement when clicking into the images for triangulation, hold down the SHIFT+CTRL keys. Note the indication of the landmark which needs to be placed (IC, AC, PC, IH) as part of the cursor symbol. After placing all of the landmarks it is recommended saving them for future use as indicated below.
Brain Indexes
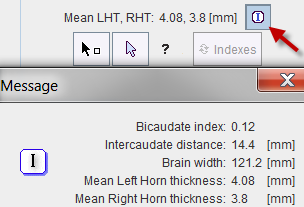
Based on the segmentation 3 indexes are calculated which will be further employed for the selection of suitable hemispheres from the knowledge base:
A summary of the indexes is shown in the user interface, and complete information can be opened with the button indicated below.
Parcellation
Deep nuclei parcellation calculates the brain structure shapes based on the knowledge base and the segmented MR image. Each hemisphere is processed individually. The user may select the Number of subjects (<=26) which are taken into account during parcellation. A higher number tends to provide more accurate results, but 8 has been found to be the optimal compromise between speed and accuracy.
Sulci parcellation uses the grey matter segment and a transformed atlas to derive cortical VOIs. The VOI Atlas to be used can be selected in the list.
Calculation is started with the Parcellation button. Its duration highly depends on the system performance (RAM, processors) and the selected number of subjects, and can range from minutes to hours.