Overview of the BFM Processing in PXMOD
In PXMOD, both the reversible and the irreversible configuration are supported. Furthermore, it is possible to allow fitting of the blood volume fraction, or to fix it at a specific value. The linear fitting is done without weighting using the singular value decomposition method.
The configurations (k4 fitted or 0, vB fitted or fixed) are specified as parameters of preprocessing. During preprocessing, the 2xN basis functions are pre-calculated and stored for pixel-wise processing. Then, the BFM analysis is applied to the TAC and the result shown for inspection. Finally, pixel-wise processing calculates maps of all parameters.
In setting up the processing for a new tracer it is recommended to enable the calculation of the a1 and a2 maps and inspect them regarding the prescribed range. If the prescribed maximum or minimum value is very frequently encountered this indicates that the range should be expanded.
Acquisition and Data Requirements
Image Data |
A dynamic PET data set. |
Blood Data |
The plasma activity from the time of injection until the end of the PET acquisition. Optionally: whole blood activity to be subtracted from the pixel-wise TACs, loaded as Whole Blood Activity in Blood Preprocessing. |
Blood Preprocessing
Data to be provided are the input curve, and optionally a whole-blood TAC for spillover correction. No whole-blood curve is defined, the input curve will also be used for spillover correction.

Model Preprocessing
During model preprocessing the basis functions are calculated for the prescribed range of a1 and a2 values. The default ranges ar a1 Î[0.0005,0.015]min-1 and a2 Î[0.06,0.6]min-1 are suitable for FDG. To change the ranges please first enable the alpha1 or alpha2 parameter and then adjust the Lower and Upper values.
Also important is the fit flag of vB and k4. If vB is checked, the blood fraction will be fitted in model preprocessing and also in the map calculation. Otherwise, the specified value will be used for spillover correction. If k4 is checked, the full 2-tissue compartment model with four parameters will be fitted in model preprocessing and map calculation. Otherwise, the specified value will be disregarded and k4 set to zero in all fits.
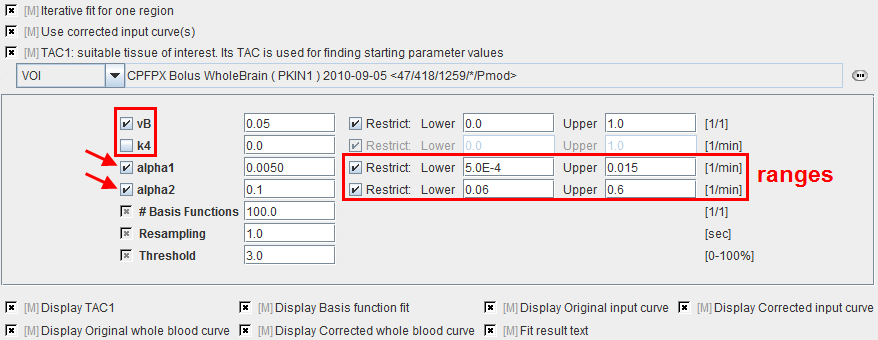
vB |
Blood volume fraction. Can be fitted or fixed. |
k4 |
Rate constant k4 in the 2-tissue compartment model. Can be fitted, otherwise it is set to 0. |
alpha1 |
First and second eigenvalue. The Lower/Upper values are used for defining the basis function ranges. |
#Basis |
Number of intermediate ai values generated between Lower and Upper. The increments are logarithmically spaced. |
Resampling |
Sampling increment applied during the basis function calculation. |
Threshold |
Discrimination threshold for background masking. All pixels with energy below Threshold[%] of the maximal energy will be masked to zero. |
Map Parameters
The example below shows a typical configuration for an irreversible mode (k4 = 0 fixed)
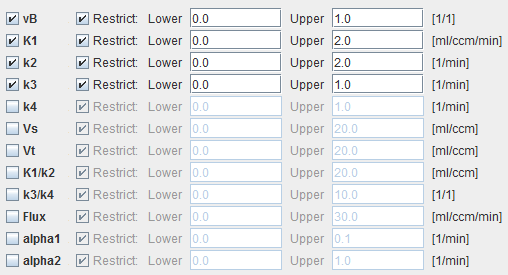
vB |
Blood volume fraction defining the pixel-wise blood spillover correction. To fit, please activate in the Preprocessing tab. |
K1,k2,k3 |
Rate constants of the 2-tissue compartment model. |
k4 |
Rate constant of the 2-tissue compartment model. A map can only be obtained if k4 has been checked for fitting in the preprocessing configuration. Otherwise the map will be zero. |
Vs |
Distribution volume of the second compartment. It is only defined for a reversible configuration where k4 has been checked for fitting. |
Vt |
Distribution volume. |
K1/k2 |
Distribution volume of the non-displaceable compartment. |
k3/k4 |
Binding potential of receptor tracers. |
Flux |
Influx of the tracer, also called Ki. |
alpha1 |
Shows the a1 and a2 values of the found solution. These values can be used to check whether the defined range was adequate. |